Revealing the ‘sweet secrets’ of coronavirus cell entry
Researchers from the Utrecht University have uncovered a sophisticated mechanism by which coronavirus spike proteins can be activated for cell entry. The study, published today in the scientific journal Nature, used powerful microscopes and computer simulations to reveal how a tiny sugar molecule binds to a human coronavirus spike and triggers exposure of components that are required to invade the host cell. These findings provide new fundamental insights into the complex mechanisms that coronavirus may use to evade the immune system and initiate an infection.
Coronavirus spikes play a crucial role in virus attachment and entry into our cells. An in-depth understanding of these proteins is important because they are key factors for transmission between species and the main targets for neutralising antibodies. To breach the host cell and deliver the viral genome, spike proteins must switch from a closed to an open state. This exposes a receptor binding domain, which then latches onto a protein receptor required for entry. For the most notorious coronaviruses, i.e. those causing SARS, MERS and COVID19, the spikes can freely alternate between these two states. However, the spike proteins from other human and animal coronaviruses have only been visualised in the closed state. This has led to the idea that most coronavirus spike proteins might not just randomly switch between states, but that there could be specific biological cues that trigger them to open.
Coronavirus attachment may be even more sophisticated than appreciated so far
To investigate this long-standing puzzle, senior authors Raoul de Groot and Daniel Hurdiss (Section of Virology, Faculty of Veterinary Medicine, Utrecht University) focused on the spike protein of human coronavirus HKU1, which, like most spikes, has only been visualised in the closed state. Collectively, the four common cold coronaviruses (HKU1, OC43, NL63 and 229E) are estimated to cause fifteen to thirty percent of respiratory tract infections each year. Previous research from the lab demonstrated that the HKU1 spike protein critically depends on binding to a specific sugar molecule, but the reason for this remained unknown. In the present study the authors discovered that sugar binding induces opening of the spike protein and exposure of the receptor binding domain, required for subsequent entry steps. The Utrecht researchers therefore found a biomolecular mechanism that was hitherto unknown to science. "It's a fine-tuned sugar switch”, says Hurdiss. “From the viruses’ perspective, it’s a clever way of keeping your Achilles heel, the receptor binding domain, hidden until the most opportune moment. Our findings paint a more elaborate picture of coronavirus attachment, with possibilities of dual receptor usage as a means of immune escape.”
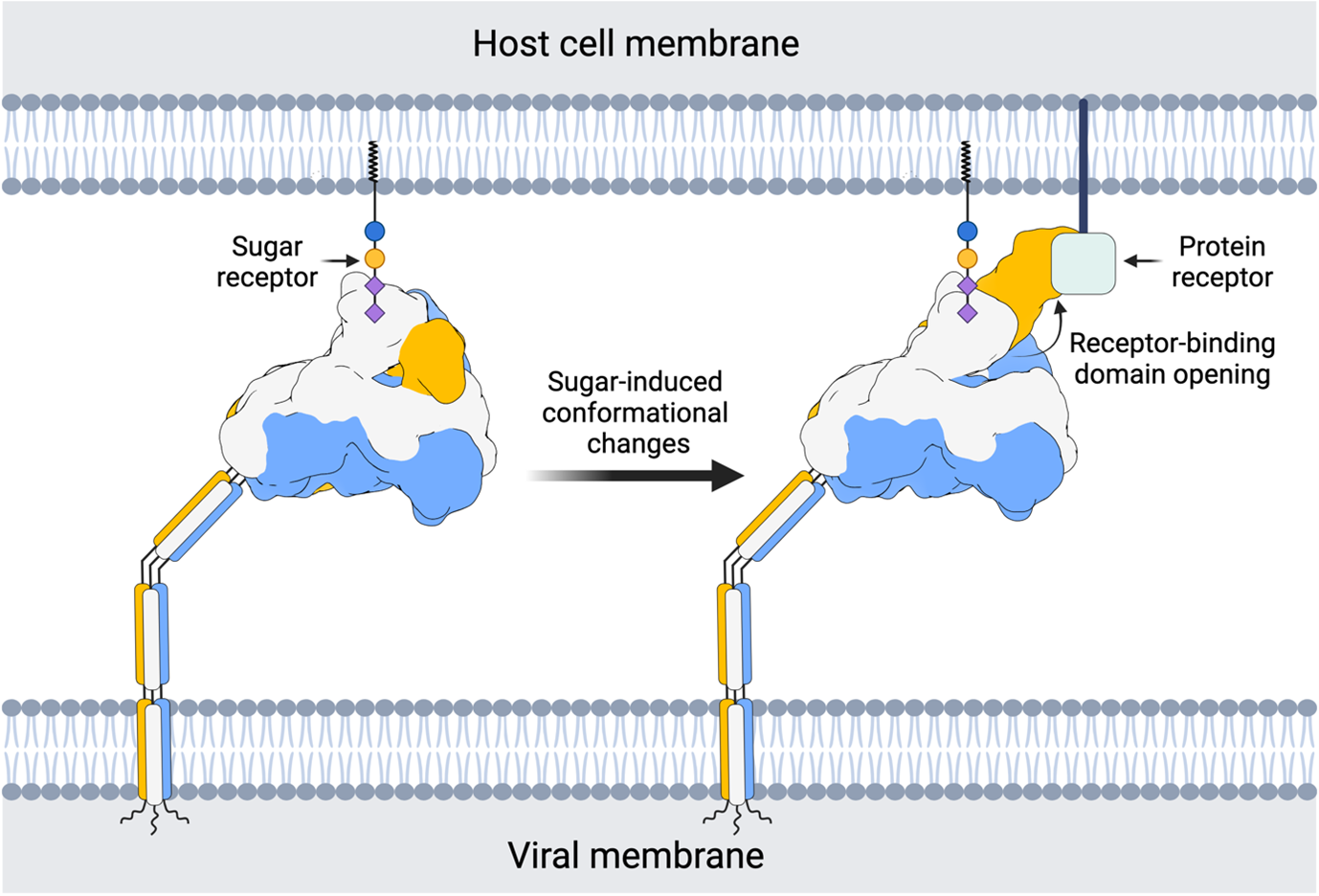
Cascade of conformational changes
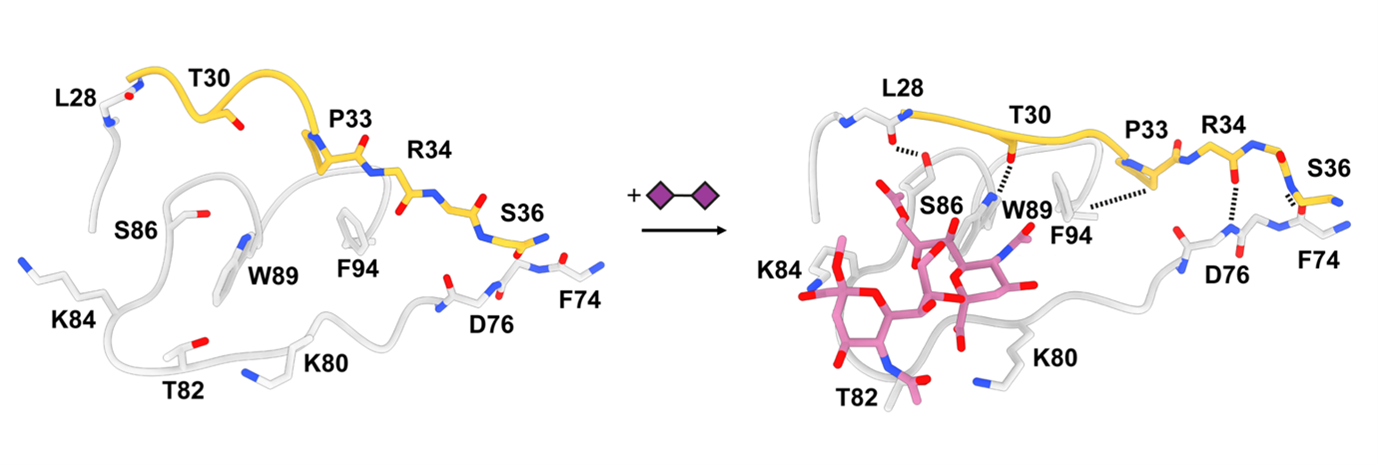
Like piecing together a movie reel from individual frames, Matti Pronker (Faculty of Science, Utrecht University and first author of the study) modelled each conformation of the spike, and deduced a stepwise series of events induced by sugar binding. Working together with Robert Creutznacher (Faculty of Veterinary Medicine, Utrecht University), and Martin Frank, computational chemist from Biognos AB, they were able to provide an atomistic explanation for how a tiny sugar molecule causes a cascade of conformational changes in this comparatively giant macromolecular complex. “It’s like a butterfly effect”, says Pronker. “It was intriguing to see how small and local conformational changes, induced by the sugar binding, trigger larger domain movements and eventually spike opening.”
Future work
“Years of research into coronavirus-sugar interactions have culminated in this work”, says De Groot. “This has very much been a joint multidisciplinary effort, profiting from the unique expertise of Geert-Jan Boons and Martin Frank, and with Yifei Lang, Zeshi Li and Ruben Hulswit laying the foundation of the present study. Our findings suggest the possibility of coronaviruses relying on multi-receptor usage with binding to sugar- or protein-based priming receptors serving as a biological cue to activate the spikes for subsequent attachment and entry steps. At the virology group, we will continue to investigate how coronaviruses bind to and enter their host cells. Such studies are of importance for understanding virus-host interactions and zoonotic transmission as well as for the development of effective countermeasures.”

