New method for zooming in on the genome’s gatekeeper
Publication in Angewandte Chemie
Researchers at Utrecht University have developed a widely applicable method to obtain insight into the interactions between proteins and chromatin at the atomic level. This is the complex of DNA and proteins in the cell that regulates everything pertaining to DNA, both during the development of the cell and over the course of its life. The method is new in that it also provides insight into the effects of the interaction on the local motions of the protein and the chromatin. “We still don’t know much about how this fundamental layer of biology works, because it is such a complex system”, according to Utrecht University research leader Dr. Hugo van Ingen. The method was published in the scientific journal Angewandte Chemie.
Chromatin acts as the gatekeeper for the genome: it either promotes or refuses access to the DNA, and therefore controls the processes in the cell. Regulatory proteins that bind to a specific location on the chromatin, influence the gatekeeper and thereby for example either activate or deactivate a gene. “The field of pharmaceutical research is interested in these kinds of interactions, because they can guide the development of new medications. However, we know only for a few proteins what the interaction with chromatin actually looks like”, Van Ingen explains.
NMR fingerprint of nucleosomes
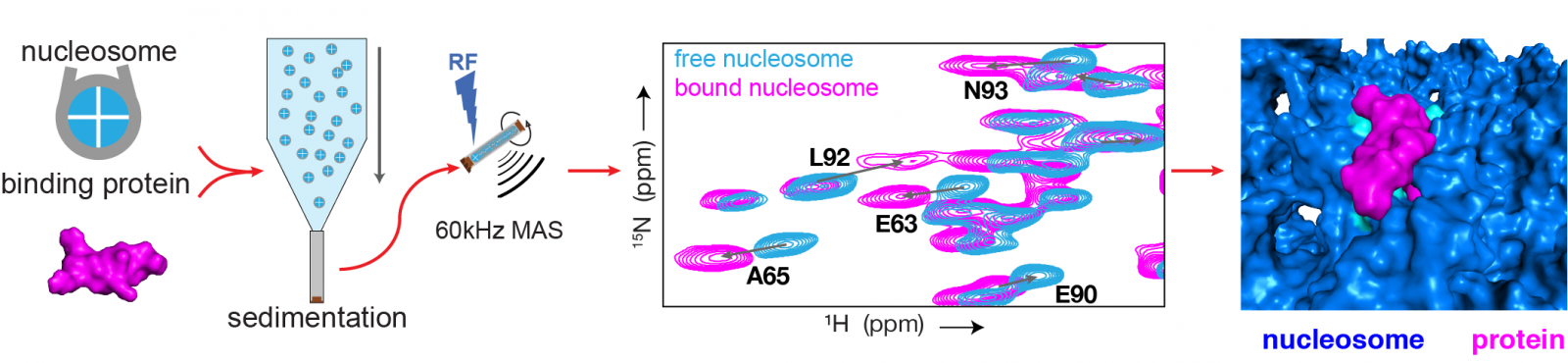
The researchers use their new method to zoom in on the individual basic units of chromatin, called nucleosomes. These are mixed in a solution together with the protein to be studied such that they bind together. Using a centrifuge, the resulting nucleosome-protein complexes are then compressed into an extremely compact sample, which is similar to the chromatin in the cell. For comparison, a sample is also created using nucleosomes without the protein. A ‘fingerprint’ of the molecule is then generated using NMR spectroscopy, a kind of MRI scanner for molecules. The differences between the two fingerprints show how the protein is bound to the nucleosome.
Real research can begin

As proof-of-principle for this technique, the researchers from Utrecht University used a viral protein for which the binding to the nucleosome is already well-understood. Now they can begin studying the many protein interactions about which little to nothing is known at the moment. “Our method places minimal demands on the proteins to be studied, so we expect that it will have many applications. Plus, it’s relatively quick, now that the difficult part of the research method has been completed. As a result, we can expand the research to studies of strings of nucleosomes, to use as models for the chromatin fibres in the cell.”
Pioneer factors
In addition to studying chromatin interactions, Van Ingen hopes to be able to combine the new method and NMR studies in a solution in order to zoom in on the specialisation of stem cells. This will also begin with the binding of between specific proteins, called pioneer factors, to nucleosomes. These interactions determine whether a stem cell develops into specific cells, such as a liver cell, a muscle cell, or a heart cell. A better understanding of this process could lead to improved methods for creating stem cells from ordinary cells.
Research method and results
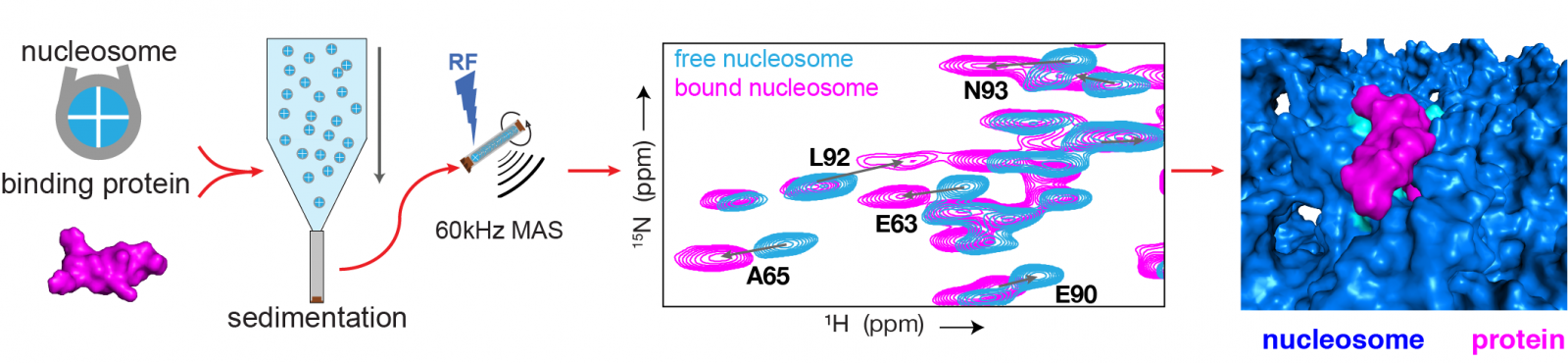
Schematic diagram of the research method (left) and an example of the results (right). (1) First, samples are created of the nucleosome-protein complexes, and of nucleosome-only complexes. (2) Both are studied using state-of-the-art NMR techniques. (3) This produces the ‘fingerprint’ spectrum: the spectrum in blue corresponds to the nucleosome without the protein, and the pink spectrum shows the nucleosome with the protein bound. Each spot on the spectrum indicates a peak, and corresponds to a single amino acid in the protein. The high resolution of the spectrum makes it possible to determine the differences between nucleosomes with- or without a protein for each peak. (4) These differences are then used to determine how exactly the protein binds to the nucleosome.
More information
This research was conducted within the research theme Science for Life, part of Utrecht University’s interdisciplinary research programme Life Sciences.
This research was a collaborative effort by the groups led by Dr. Hugo van Ingen (fluid NMR Spectroscopy) and Prof. Marc Baldus (solid NMR Spectroscopy), both affiliated with Utrecht University. This research was funded in part by NWO (Vidi grant for Hugo van Ingen).
Video ‘What is NMR?’
Publication
Site‐Specific Studies of Nucleosome Interactions by Solid‐State NMR Spectroscopy
Dr. ShengQi Xiang#, Ulric B. le Paige#, Velten Horn, Dr. Klaartje Houben, Prof. Marc Baldus, Dr. Hugo van Ingen: all authors are affiliated with Utrecht University
Angewandte Chemie, March 2018, https://doi.org/10.1002/anie.201713158
# equal contributions
Science for Life
This research was conducted within the research theme Science for Life, part of Utrecht University’s interdisciplinary research programme Life Sciences.

