Exposing new vulnerabilities on the tumor surface
Publication in Nature Communications
Cancer tumors are a diverse collection of cells. Researchers from Utrecht University, Hubrecht Institute and UMC Utrecht discovered that the tumor cells of one patient also differ in its surface antigens. “Such antigen targets are very interesting to pursue for personalized immunotherapy”, Wei Wu says. The researchers publish their findings in Nature Communications.
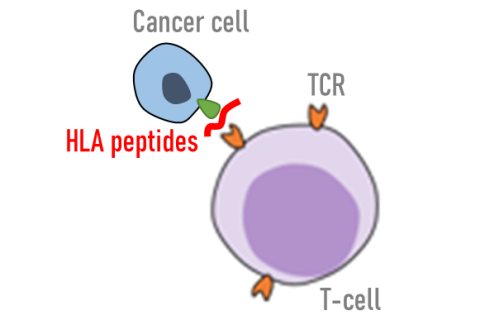
All cells in the body are scanned constantly by our own immune system. Healthy cells present small fragments of degraded proteins, as ‘self’ HLA peptide antigens, that are well-tolerated by our individual immune systems. Aberrant and cancerous cells, on the other hand, may present unique or mutated surface marks that can trigger T-cells to become activated.
This surveillance through HLA peptide antigens forms the basis to trigger and modulate T-cell responses. Hence HLA peptide presentation can influence tumor clearance, and can also be exploited in tumor immunotherapy and vaccination. Here, researchers of Utrecht University, the UMCU and the Hubrecht Institute investigated clonal diversity in tumor-specific HLA peptide presentation, for fundamental rules to target tumors.
Tumor heterogeneity as major cause of therapeutic resistance
Tumors are not uniform, but a diverse collection of cells; some of these may resist treatment better than others and can survive toxic stress of chemical drugs to re-establish during cancer relapse. For many chemical drugs, this model of tumor evolution acting on intrinsic tumor heterogeneity is well-documented. It is not known, however, if HLA peptide presentation is also heterogenous. This knowledge will potentially help us to understand escape from immunosurveillance and from immunotherapy.

“A key question to address is whether tumor heterogeneity is also reflected in the antigens presented by tumor cells. This is an important question, because if such heterogeneity exists, some tumor cells can still evade the immune system just like how drug-resistant cells evade regular therapy”, says Laura Demmers, PhD candidate at the Biomolecular Mass Spectrometry and Proteomics group.
Integrating technologies for deeper characterization of tumor surface presentation
This work required the initial tumor and normal single cells, from one colorectal cancer patient, to be preserved and amplified using the organoid platform, in collaboration with Hans Clevers at the Hubrecht Institute. This gave a boost to the detection of unique tumor HLA peptide antigens by alleviating the bottleneck of patient material scarcity.
HLA peptide antigens may be smaller fragments than proteins, but these short peptides are in no ways easier to characterize than whole-proteins. Such peptides from tumor cells may also harbor protein-coding mutations that further complicate their identification and the inference of their origin. To solve this, patient and tumor clone-specific sequencing was also performed in collaboration with Edwin Cuppen at the UMC Utrecht. The fundamental detection of HLA peptide antigens was further boosted by the complementary use of two fragmentation methods in the mass spectrometer, to capitalize on different chemical properties of diverse HLA peptide species.
This collaborative work between the researchers from the UMCU, UU and Hubrecht Institute was supported by the X-Omics National Roadmap Initiative and the Utrecht Molecular Immunology Hub and the Gravitation Program Institute for Chemical Immunology.
The tumor-specific HLA peptide antigen landscape
The team at Utrecht University found that about 300 HLA peptide antigens were presented exclusively by the tumor cells. Many of these antigens originate from notorious oncogenic proteins and are shared amongst all tumor clones analysed. Such antigen targets are very interesting to pursue for personalized immunotherapy, and in theory should be equally effective against all tumor cells of the same patient.
More intriguingly, the HLA peptide repertoire between different tumor clones of the same individual also differed significantly by 15-25%, as the data suggested. Such variations in antigen presentation may be sufficient to drive immunotherapeutic escape, in mechanisms similar to the selection of resistant phenotypes in chemotherapy. To overcome this, targeting multiple antigens simultaneously may help, but better solutions may emerge while re-tracing the mechanistic path in HLA peptide antigen presentation.
While sieving through the antigens consistently presented only by tumor cells, the team found quite some fragments of DNA damage sensing and repair proteins, for instance, peptides from BRCA proteins. This made a lot sense in tumors, since getting rid of DNA repair mechanisms would allow accumulation of deleterious traits towards malignancy.

“The key to treat cancer lies in the jab we take at the Achilles’ heel”, says Wei Wu, Assistant Professor at the Department of Pharmaceutical Sciences, “while keeping DNA repair silent provides a huge advantage for tumors to progress, it inevitably exposes a weakness on the cell surface, that we can target. The same may also apply to other cancers, since loss of DNA repair is also conserved across many other cancers”.

“An aspect of the work I also like is that we use the organoid technology to actually amplify the signal of a single tumor cell, providing an alternative route for single cell proteomics”, says Albert Heck, Professor of Chemistry and Pharmaceutical Sciences at Utrecht University.
This work takes on a new angle to perceive the ideal choice of cancer neoantigens. Instead of focusing on over-expressed oncogenic and mutated proteins, the authors propose that peptide fragments from the degradation of tumor suppressor proteins may also be relevant candidates.
Publication
Single-cell derived tumor organoids display diversity in HLA class I peptide presentation Nature Communications, 21 October 2020. Laura C. Demmers*, Kai Kretzschmar, Arne Van Hoeck, Yotam E. Bar-Epraïm, Henk W. P. van den Toorn*, Mandy Koomen, Gijs van Son, Joost van Gorp, Apollo Pronk, Niels Smakman, Edwin Cuppen, Hans Clevers*, Albert J. R. Heck* & Wei Wu*
*Affiliated with Utrecht University

