Highlights
This page archives our highlights that have reached the news. Appearances in national newspapers and radio/TV can be found in the left menu. A pdf with information about our institute and all UU news items of our institute between the Symposium of October 2016 and October 2018 can be downloaded here. The following four showcases give a sample of our research.
Covid-19 showcases
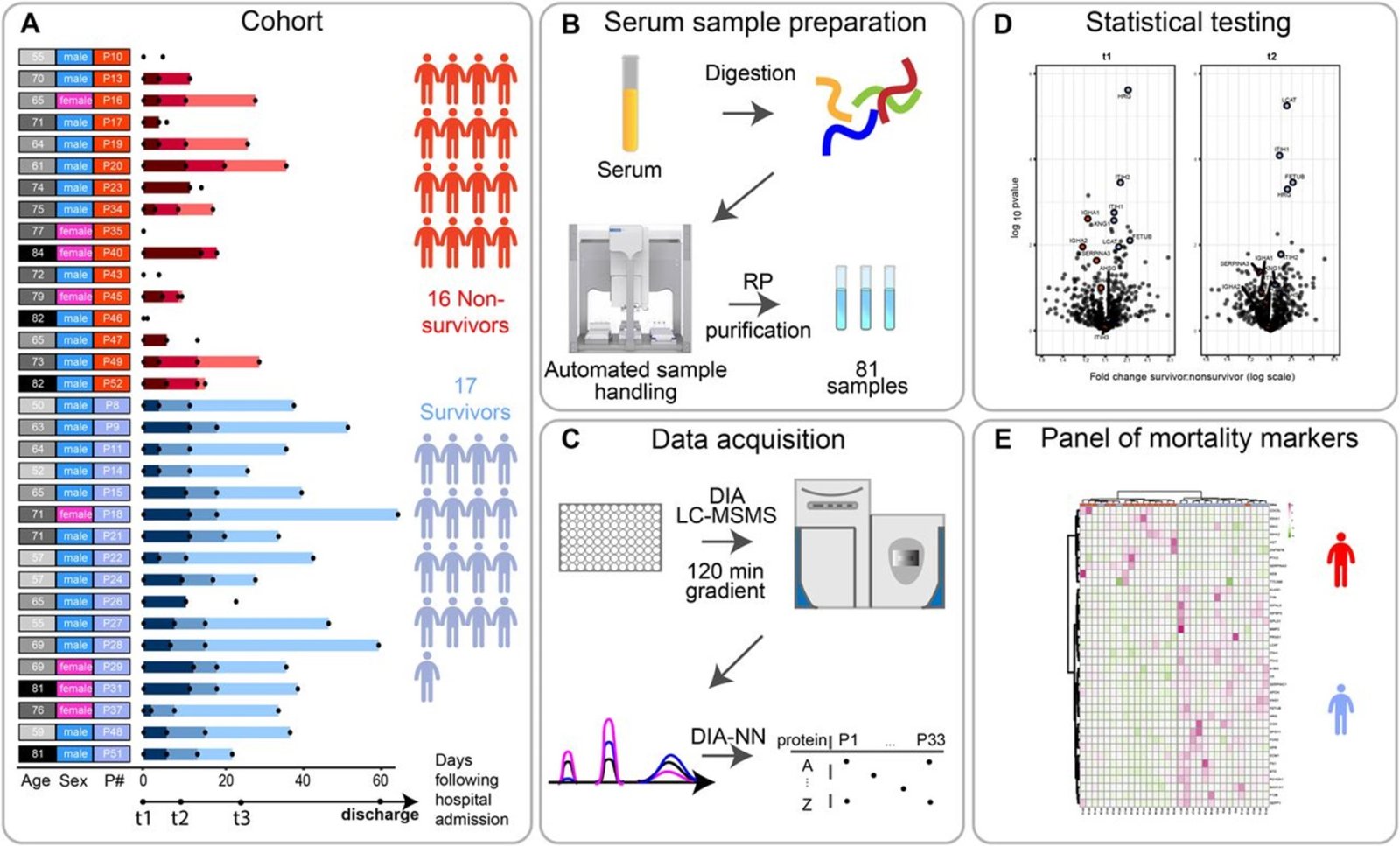
With the advent of Covid19 pandemic, and the unprecedented lockdowns, society realised its vulnerability. Fortunately, the scientific response was quick with PCR tests, antigen selftests, five vaccines and three drugs. Also our Divisions were able to focus their technical expertise on Covid19, leading to published studies, listed here and vaccine research listed here. With machine learning our new staff member dr. Alejandro Lopez Rincon designed specific primer sequences for PCR (1). The toxicity of the repurposed drug chloroquine was studied (2). The attachment receptors of the virion were studied with glycan arrays, discovering sialic acids and heparan sulfate as entry points (3). Protective antibodies were found in breast milk (4). Serum proteomes were measured and linked to disease severity (5, Figure 1). The vaccine (thermal) stability was studied (6). Some vaccine adverse effects were studied for the EMA (7). The impact of the pandemic on pharmaceutical care as well as on wellbeing was studied (8). Our staff member prof. de Boer, chair of the Medicines Evaluation Board, was actively involved with the regulatory issues, as well as with informing the general public on national television.
- Lopez-Rincon A, Tonda A, Mendoza-Maldonado L, Mulders DGJC, Molenkamp R, Perez-Romero CA, Claassen E, Garssen J, Kraneveld AD. Classification and specific primer design for accurate detection of SARS-CoV-2 using deep learning, Sci Rep. 2021 Jan 13;11(1):947. doi: 10.1038/s41598-020-80363-5.
- van den Broek MPH, Möhlmann JE, Abeln BGS, Liebregts M, van Dijk VF, van de Garde EMW. Chloroquine- induced QTc prolongation in COVID-19 patients. Neth Heart J. 2020 Jul;28(7-8):406-409. doi: 10.1007/ s12471-020-01429-7.
- Nguyen L, McCord KA, Bui DT, Bouwman KM, Kitova EN, Elaish M, Kumawat D, Daskhan GC, Tomris I, Han L, Chopra P, Yang TJ, Willows SD, Mason AL, Mahal LK, Lowary TL, West LJ, Hsu SD, Hobman T, Tompkins SM, Boons GJ, de Vries RP, Macauley MS, Klassen JS. Sialic acid-containing glycolipids mediate binding and viral entry of SARS-CoV-2. Nat Chem Biol. 2022 Jan;18(1):81-90. doi: 10.1038/s41589-021-00924-1.
- van Keulen BJ, Romijn M, Bondt A, Dingess KA, Kontopodi E, van der Straten K, den Boer MA, Burger JA, Poniman M, Bosch BJ, Brouwer PJM, de Groot CJM, Hoek M, Li W, Pajkrt D, Sanders RW, Schoonderwoerd A, Tamara S, Timmermans RAH, Vidarsson G, Stittelaar KJ, Rispens TT, Hettinga KA, van Gils MJ, Heck AJR, van Goudoever JB. Human Milk from Previously COVID-19-Infected Mothers: The Effect of Pasteurization on Specific Antibodies and Neutralization Capacity. Nutrients. 2021 May 13;13(5):1645. doi: 10.3390/nu13051645.
- Völlmy F, van den Toorn H, Zenezini Chiozzi R, Zucchetti O, Papi A, Volta CA, Marracino L, Vieceli Dalla Sega F, Fortini F, Demichev V, Tober-Lau P, Campo G, Contoli M, Ralser M, Kurth F, Spadaro S, Rizzo P, Heck AJ. A serum proteome signature to predict mortality in severe COVID-19 patients. Life Sci Alliance. 2021 Jul 5;4(9):e202101099. doi: 10.26508/lsa.202101099.
- Crommelin DJA, Anchordoquy TJ, Volkin DB, Jiskoot W, Mastrobattista E. Addressing the Cold Reality of mRNA Vaccine Stability. J Pharm Sci. 2021 Mar;110(3):997-1001. doi: 10.1016/j.xphs.2020.12.006. Epub 2020 Dec 13.
- Sturkenboom, MCJM, Messina, D, Paoletti, O, de Burgos, A, Garcia, P, Huerta Álvarez Consuelo, Llorente, A, Klungel, O, Martin, M, Martinez, M, Martin, I, Overbeek, J, Souverein, P, Swart, K, & Gini, R. (2022). Cohort monitoring of Adverse Events of Special Interest and COVID-19 diagnoses prior to and after COVID-19 vaccination (1.0). Zenodo. https://doi.org/10.5281/zenodo.6762311
- Koster ES, Philbert D, Bouvy ML. Impact of the COVID-19 epidemic on the provision of pharmaceutical care in community pharmacies. Res Social Adm Pharm. 2021 Jan;17(1):2002-2004. doi: 10.1016/j. sapharm.2020.07.001. Epub 2020 Jul 2.
ATMP showcases
A common research theme within UIPS is on advanced therapies, which include gene and cell therapies. Several initiatives and collaborations are ongoing, of which a few are listed below.
Cell-based therapies
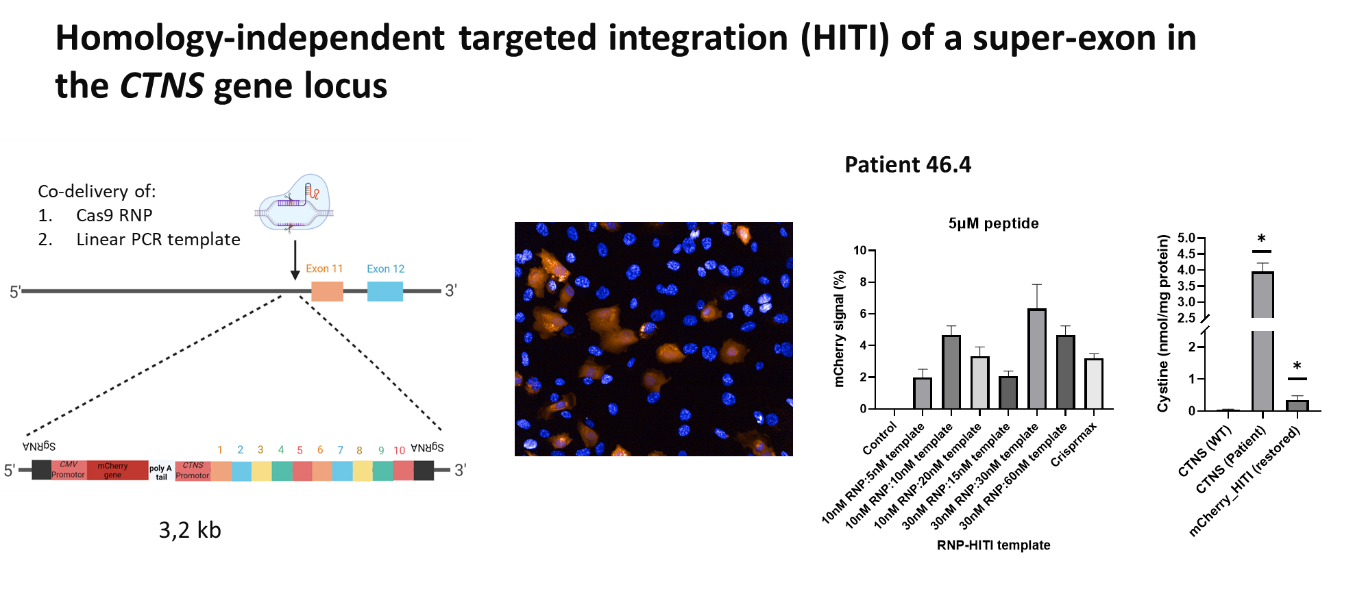
In a collaborative project between the Pharmacology group (Masereeuw/Janssen/Sendino-Garvi) and the Pharmaceutics group (Mastrobattista, de Jong, Oktem), we have worked on gene editing strategies to treat nephropathic cystinosis, a rare monogenic kidney disease caused by mutations in the CTNS gene leading to toxic cystine accumulation in cells (1). Making use of a peptide-based delivery system developed in the Pharmaceutics group, CRISPR Cas9 ribonucleoprotein complexes and a DNA template could be efficiently delivered into the patient cells, resulting in homology-independent gene insertion of the therapeutic DNA. With this approach, we were able to correct cystinosis patient-derived fibroblasts, leading to restoration of normal intracellular cystine levels (unpublished data below). This method could in principle by used for autologous cell therapy in which the hematopoietic stem cells of a patient will be ex vivo corrected, before reinfusion into the patient. Besides, it could be used to construct advanced in vitro (diseased) kidney models. See figure 2.
In another project in collaboration with Glycostem B.V. and sponsored by Health Holland the Pharmaceutics group (Caiazzo/Mastrobattista) is working on the development of non-viral delivery methods to genetically modify NK cells for their NK cell therapy pipeline. Using polymeric and lipid nanoparticles, an attempt is being made to transfect mRNA into large quantities of NK cells in a safe and economical manner (2).
Gene-therapies
The Pharmaceutics group has an active collaboration with various clinical groups in the Netherlands to develop non-viral delivery methods for CRISPR-Cas (3). In a collaboration project with the Tytgat institute for liver and intestinal research (Bosma), the group is developing a lipid-based nanoparticle system to deliver CRISPR-Cas to hepatocytes to correct the monogenetic disease progressive familiar intrahepatic cholestasis type 3.
Innovation Center for Advanced Therapies (iCAT)
In a collaborative project between University Medical Center Utrecht (UMCU), the Hubrecht Institute, Princess Máxima Center for pediatric oncology, The Regenerative Medicine Centre Utrecht and the Utrecht Institute for Pharmaceutical Sciences, the aim is to accelerate the preclinical and clinical development of advanced therapies, for which an innovation center has been established (https://youtu.be/DcRkAkYE1I8). Within iCAT several research groups of the department of Pharmaceutical Sciences are actively involved, including the Pharmacology group (advanced in vitro models/regenerative medicine), the Pharmaceutics group (GMP production of synthetic gene delivery vectors) and the Pharmacoepidemiology and Clinical Pharmacy group (health technology assessment of ATMPs). Within iCAT, new research insights can be translated into new therapies and diagnostic research models by researchers in collaboration with companies. See Figure 3.
- Jamalpoor, A. et al. (2021) Molecular Mechanisms and Treatment Options of Nephropathic Cystinosis. Trends in Molecular Medicine, July 2021, Vol. 27, No. 7
- Lou, B., Lau, C. Y. J., Hennink, W. E. & Mastrobattista, E. Preparation of mRNA Polyplexes with Post-conjugated Endosome-Disruptive Peptides. Methods Mol. Biol. 2355, 275–286 (2021)
- Walther, J. et al. Impact of Formulation Conditions on Lipid Nanoparticle Characteristics and Functional Delivery of CRISPR RNP for Gene Knock-Out and Correction. Pharmaceutics 14, 213 (2022)
Regenerative Pharmaceutical Sciences Showcases
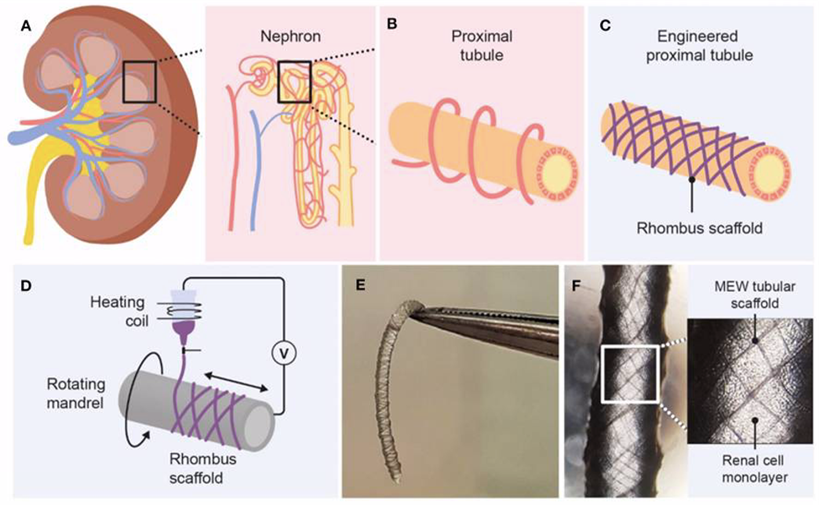
Within Utrecht Life Sciences domain Regenerative Medicine & Stem Cells novel therapeutic strategies are being developed by supporting self-repair of injured tissues as well as tissue function replacements with a final goal to improve the quality of life of patients with (chronic) severe disorders. Research at UIPS adheres to these aims by multidisciplinary approaches in developing innovative technologies such as 3-dimensional advanced tissue cultures with microfluidics (organs-on-chip technology), biofabrication, biomaterials, cell-based therapies and ethical and regulatory aspects related to the engineering of tissues and organs. The Vermonden lab, in close collaboration with the Masereeuw lab and the Malda Lab (UMCU/UU Veterinary Medicine), have developed unique shrinking biomaterials to approach physiological tissue dimensions with bio-printing (1). Through the identification of chemical design parameters that affect the shrinking properties combined with insights in cell-material interactions, innovative next generation materials for 3D bioprinting and tissue engineering applications are being developed. One of the most challenging organs is the kidney. Through surface topographical guiding by printed scaffolds, kidney cells can be steered to form proximal tubules in vitro (2). These functional units can be used for drug screening, may act as alternatives for animal experiments in drug development and applied in bioartificial kidneys for kidney replacement therapies. By using human adult stem cell derived kidney tubular organoids (tubuloids) (3), therapies can be personalized advancing the possibilities of clinical implementation. Such application can be considered an Advanced Therapy Medicinal Product (ATMP), which requires a challenging alternative regulatory trajectory (4).
- Gong J, Schuurmans CCL, Genderen AMV, Cao X, Li W, Cheng F, He JJ, López A, Huerta V, Manríquez J, Li R, Li H, Delavaux C, Sebastian S, Capendale PE, Wang H, Xie J, Yu M, Masereeuw R, Vermonden T, Zhang YS. Complexation-induced resolution enhancement of 3D-printed hydrogel constructs. Nature Commun. 2020; 11(1):1267.
- van Genderen AM, Jansen K, Kristen M, van Duijn J, Li Y, Schuurmans CCL, Malda J, Vermonden T, Jansen J, Masereeuw R, Castilho M. Topographic Guidance in Melt-Electrowritten Tubular Scaffolds Enhances Engineered Kidney Tubule Performance. Frontiers Bioeng Biotechnol. 2021;8: 617364
- Schutgens F, Rookmaaker MB, Margaritis T, Rios A, Ammerlaan C, Jansen J, Gijzen L, Vormann M, Vonk A, Viveen M, Yengej FY, Derakhshan S, de Winter-de Groot KM, Artegiani B, van Boxtel R, Cuppen E, Hendrickx APA, van den Heuvel-Eibrink MM, Heitzer E, Lanz H, Beekman J, Murk JL, Masereeuw R, Holstege F, Drost J, Verhaar MC, Clevers H. Tubuloids derived from human adult kidney and urine for personalized disease modeling. Nature Biotechnol. 2019; 37(3):303-313.
- Ten Ham RMT, Hoekman J, Hövels AM, Broekmans AW, Leufkens HGM, Klungel OH. Challenges in Advanced Therapy Medicinal Product Development: A Survey among Companies in Europe. Mol Ther Methods Clin Dev. 2018; 11:121-130.
Glycans Showcases
In the period of review, we have established expertise in glycobiology in the Divisions CBDD, PCOL and BMSP. Prof. Boons, prof. Pieters, dr. Wennekes and dr. Jongkees have transformed the medicinal chemistry group into a chemical glycobiology and drug discovery group. The successes depend for a large part on the biological and immunological expertise of dr. de Vries and dr. Heesters. Dr. Stahl from Danone Nutricia adds his expertise on glycan analysis and is a liaison between CBDD, BMSP and PCOL. At BMSP dr. Reiding, dr. Snijder and prof. Heck study glycoproteins by MS. And prof. Garssen, working in PCOL and Danone Nutricia has expertise of glycans and immunology. A research example, also showing the collaboration between institutes, can be found in this case presented by the Bijvoet centre:
Glycan receptors of human respiratory viruses (with Bijvoet Center)
Numerous viruses initiate infection by binding to cell surface glycans of the host. The selectivities of viral receptor binding proteins for specific glycan structures critically determine host range, targeted tissues and cells, and pathogenesis. Viral proteins are generally also glycosylated which can modulate these interactions. The Boons lab (Bijvoet and UIPS) in collaboration with the De Groot lab (UU Veterinary Medicine) have developed chemoenzymatic synthetic methods that can provide almost every naturally occurring sialoglycan receptor having different linkages and hydroxyl modifications. The approach was used to develop a glycan microarray to determine receptor specificities of human and animal viruses that engage with O-acetylated sialic acids including betacoronaviruses, toroviruses and influenza C and D viruses. It uncovered host-specific patterns of receptor recognition relating to both the O-acetylation pattern and sialic acid glycosidic linkage type. Our data revealed that viruses adapted to humans underwent convergent evolution to become highly selective for O-acetylated forms of α2,8-linked disialylated structures commonly found on glycolipids. Further, the Snijder lab (Bijvoet) in collaboration with the Hurdiss and De Groot labs (UU Veterinary Medicine) have used an integrated structural virology approach, combining cryoEM and mass spectrometry to map the glycans of the viral envelope glycoproteins to unprecedented detail. It provided insight into the evolution and host adaptation of human betacoronaviruses, and demonstrate the utility of cryo-EM for studying small, heavily glycosylated proteins. See Figure 4.
- Li, Zeshi, Yifei Lang, Lin Liu, Mehman I. Bunyatov, Angelic Isaza Sarmiento, Raoul J. de Groot, and Geert-Jan Boons. “Synthetic O-acetylated sialosides facilitate functional receptor identification for human respiratory viruses.” Nature Chemistry 13: 496-503 (2021).
- Hurdiss, Daniel L., Ieva Drulyte, Yifei Lang, Tatiana M. Shamorkina, Matti F. Pronker, Frank JM van Kuppeveld, Joost Snijder, and Raoul J. de Groot. “Cryo-EM structure of coronavirus-HKU1 haemagglutinin esterase reveals architectural changes arising from prolonged circulation in humans.” Nature Comm. 11: 1-10 (2020).
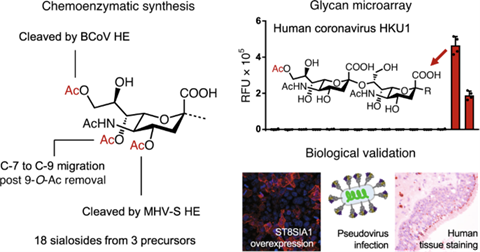
Figure 4. An unprecedented collection of O-acetylated sialoglycans was prepared by diversification of common synthetic precursors by regioselective de-O-acetylation by coronaviral hemagglutinin-esterase (HE) combined with C-7 to C-9 acetyl ester migration. The resulting compound library was printed on streptavidin-coated glass slides to give a microarray to investigate receptor binding specificities of viral envelope glycoproteins, including spike proteins and HEs from animal and human coronaviruses. It showed host-specific patterns of receptor recognition and revealed that three distinct human respiratory viruses selectively bind 9-O-acetylated α2,8-linked disialosides. Immunofluorescence and cell entry studies demonstrated that such glycotope as part of gangliosides, are functional receptors for human coronavirus.





