BSc/MSc Internships Van Ingen group
Structural basis of epigenetics & chromatin function
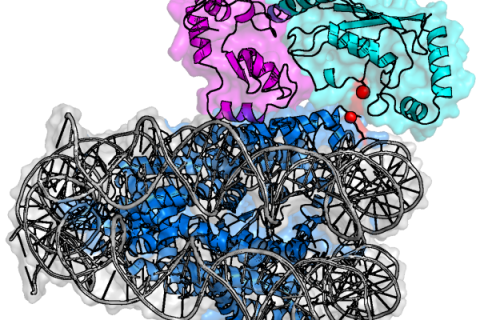
The Van Ingen group offers several possibilities for BSc or MSc internships. Do not hesitate to contact Hugo (h.vaningen_AT_uu.nl) for up-to-date possibilities!
Research Context Our research mainly focusses on the structural basis of epigenetics: how do proteins operate on nucleosomes to control DNA repair or gene regulation? These are fundamental questions that are crucial for our understanding of how cells function and may lead to new directions to treat disease.
Research Highlight For example, we recently elucidated how an E3/E2 enzyme pair recognizes the nucleosome and can specifically ubiquitinate histone H2A during the DNA damage response. By integrating NMR, cross-linking and biochemical data, we could determine the structure of this complex. We showed how one mutation in the histones can selectively interfere with binding of this E3. This mutation was later shown to be one of the most occuring histone mutations in some cancers, highlight how these cells actively alter the DNA damage response.
Internship Projects Projects are usually tied to on-going research of a PhD student, e.g. on nucleosome remodellers, the molecular machines that are capable of moving nucleosomes around, histone variants and histone chaperones. Typically, projects entail a combination of biochemistry, NMR and computational analysis. Depending on your interest projects may focus more on one of these aspects. In particular for MSc internship we aim to include your data in a publication.
What you will do Typically students will learn to do a number of the following: histone and protein expression and purification; isotope labeling; histone refolding; nucleosome reconstitions; production of DNA; setup of NMR experiments; analysis of NMR spectra; motional modelling; integrative modeling; activity tests; etc.
Current projects
- Dissecting motions in the ISWI remodeller
- Towards the cellular binding mode of chromatin bound proteins
- Build your own chromosome 6 nucleosome!
- Develop in-house production of Sca1
Examples of previous internships:
- biochemical characterization of nucleosomes containing a histone variant by mutagenesis and micrococcal DNA digestion
- structure determination of macrocyclical peptides by solution NMR (publication)
- residue-specific pKa determination of ionizable groups in histone proteins by solution NMR
- analysis of a weakly coupled, magnetically inequivalent spin system (publication)

