How chromosomes are bound to be separated in cell division
The first structure of the inner kinetochore bound to a centromeric nucleosome shows how chromosomes are bound before being segregated into two new cells
When our cells divide, it is important that the pairs of chromosomes are correctly segregated, as errors in this process cause serious problems. For over a century, kinetochores have been recognised as the critical cellular structures responsible for attaching the chromosomes to the microtubules that direct this chromosomal segregation. However, how exactly kinetochores recognise the centromere, the central point that links the two halves of a chromosome, has been a long-standing question. David Barford’s group in the LMB’s Structural Studies Division and Stephen McLaughlin of the LMB’s Biophysics Facility, have now determined a structure of a kinetochore complex that suggests the answer to this question.
Kinetochores
Kinetochores are large protein complexes formed of two domains: the inner and outer kinetochore. The inner kinetochore consists of a complex of 14 subunits, known as CCAN. Our DNA is standardly wrapped up tightly around a set of histone proteins known as a nucleosome. At centromeres, one of the standard histone proteins is replaced by a protein called Cenp-A to generate a specialised Cenp-A nucleosome. CCAN is known to interact with the Cenp-A nucleosome and form a bridge to the outer kinetochore. However, structures of only a few CCAN subunits have been solved and it is not known how they are organised around each other in CCAN or how CCAN binds Cenp-A nucleosomes.
To investigate this question, Kaige Yan, Jing Yang, and Ziguo Zhang from David’s group produced and purified the subunits of the yeast inner kinetochore complex from insect cells, assembled CCAN bound to a Cenp-A-containing nucleosome, and used electron cryo-microscopy to reveal the structure of the complex. A unique feature of Cenp-A nucleosomes is that the DNA coiled around it is unwrapped by 20 base-pairs at each end. This new structure shows how the unwrapped DNA provides the major interaction interface between CCAN and the Cenp-A nucleosome. As this feature is unique to Cenp-A nucleosomes, this shows why kinetochores specifically assemble at centromeres.
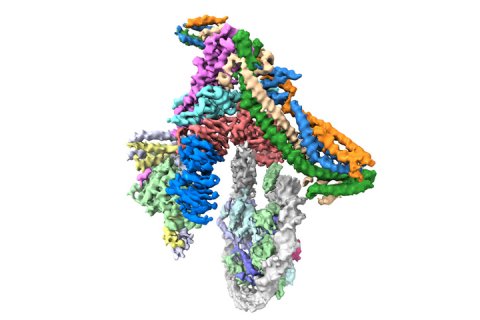
The cryo-EM structure was validated using cross-linking mass spectrometry in collaboration with Albert Heck’s group at Utrecht University.
The model for Cenp-A nucleosome binding was tested in vivo using mutant yeast strains developed by Ann Ehrenhofer-Murray at Humboldt University of Berlin.
Although there are differences in scale between the systems controlling chromosome segregation in yeast and vertebrates, the conservation of the constituent components suggests that the mechanism of recognition of centromeres by kinetochores identified here would be similar in our own cells. Understanding how this process is controlled is important as errors in cell division underlie cancers and developmental defects. This new structure provides a framework for understanding decades of biochemical and genetic studies on the centromere and kinetochore, and serves as a basis for future mechanistic studies.
The work was funded by the MRC, CRUK, European Commission, and the German Research Foundation.
Publication
Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome. Yan, K., Yang, J., Zhang, Z., McLaughlin, SH., Chang, L., Fasci, D., Ehrenhofer-Murray, AE., Heck, AJR., Barford, D., Nature (2019) Oct. 10th, DOI: 2019-01-01089C
Source:
Medical Research Council Laboratory of Molecular Biology

